Hi,
At my workplace we use the “ExtraMainDicomTags” feature from the DicomWeb plugin and also we configured the plugin to work in “ExtraMainDicomTags” mode as is described here. Unfortunately, our DICOM viewer fails when it tries to obtain some studies metadata, the error in the logs is: E1004 20:11:24.502791 OrthancException.cpp:61] Bad type for a parameter: Bad Parameter type for tag 0021,11fe
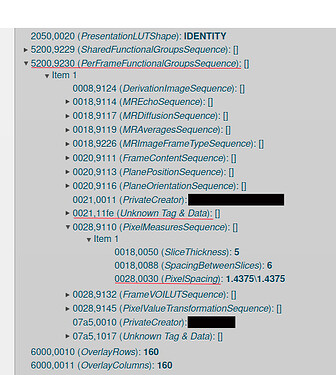
After some research on the error we found 0021,11fe it’s a private tag that is irrelevant for our viewer renderization process, but it’s stored as part of the “ExtraMainDicomTags” because it’s inside (5200,9230) PerFrameFunctionalGroupsSequence sequence, which is include on our “ExtraMainDicomTags” list. Remove PerFrameFunctionalGroupsSequence from the list and reconstruct the tags of the study solves the problem of metadata obtention and the viewer successfully render the study, but for some of our viewer features we use the (0028,0030) PixelSpacing tag that is also inside the sequence, so remove the sequence from the list is not viable for us. In fact, the only relevant tag for our viewer from that sequence is PixelSpacing but in our tests we realize that nested values are not stored in db if the container sequence is not in the list of “ExtraMainDicomTags”, which causes that values irrelevant for study rendering to be saved in the Orthanc database.
We tried to add the 0021,11fe to the Dictionary in Orthanc configuration as is suggested here and here, but the error persist. We also tried to remove all the private tags from the studies with the problem as is described here, despite we use the "RemovePrivateTags": true option in the modification request and delete the previous instance before upload the modified one, the private tags are still there.
Is there a way to store just some tags from a sequence on the Orthanc DB without store the entire sequence? In case it isn’t possible, Is there a way to ignore some tags on DicomWeb requests?
We are using Orthanc 1.12.1 and dicomWeb plugin 1.15, the script used to modify the affected studies instances can be found here, an example of our Orthanc configuration can be found here, and below is a screenshot with the reference of the tags structure of an affected instance.
Thanks in advice and best regards,
Rodrigo
Hi Rodrigo,
I’m not sure that we have tested the “RemotePrivateTags” against private tags stored inside a sequence. Could you share an anonymized file for us to add to our test suite ?
There is currently no way to tune the ExtraMainDicomTags configuration to avoid storing the Private tags inside the sequence either.
Please also note that since we have released version 1.15 of DicomWeb, we have introduced some caching for series metadata. This removes the burden of configuring the ExtraMainDicomTags and actually is at least as fast and always correct ! Could you give it a try on your system ? You would just have to reconfigure
"StudiesMetadata" : "Full",
"SeriesMetadata" : "Full"
Hope this helps,
Alain.
Hi Alain,
Sorry for the late response, it’s been a busy few weeks.
We have already tested the “Full” option of the DicomWeb plugin but although it solves the display errors, it doesn’t suit our use case because we store the instances in an object storage, which makes obtaining tags that are not are stored in the database an slow operation. The cache you mention can solve part of the problem, but our users usually view the studies only once and then they are not accessed again, so the cache has no effect in practice. So the “ExtraMainDicomsTags” option is the one that best suits our use case.
Apart from the problem with private tags, we also found problems with the (6000,0050) OverlayOrigin tag present in mammograms, which, when added to the “ExtraMainDicomsTags” causes Orthanc to display the following error: OrthancException.cpp: 61] Bad file format: While creating a DICOM instance, tag (6000,0050) has out-of-range value: "1\1".
In this link you can found an anonymized version of the study with the problem with private tags and another with the OverlayOrigin tag, so you can reproduce the problems.
We’re available to contribute a patch to solve these problems if you give us some guidance on where to start.
Thanks in advice,
Best regards
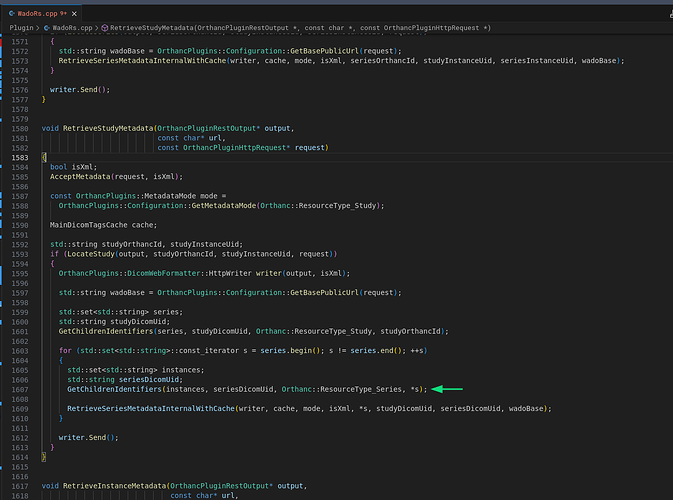
After some research on the problem of the out-of-range value, we found that the error is in some point between the value extraction from database and the cast of the value to an Signed Short (SS) representation. Also, we detect that in the DicomWeb plugin at WadoRs.cpp:1605 the variable instances is declared and the GetChildrenIdentifiers method set his value, but that cariable is not used in the rest of the code, this also duplicates the requests to the Orthanc’s internal series metadata endpoint.
Hi Rodrigo,
I still recommend using the “Full” mode even if your users are looking at the image only once. The cache is populated when the images are received (before the doctors look at them) and the overhead in terms of storage is less than 0.1%.
Sorry but I currently do not have time to dig in this issue. We also improved the “Full” mode to ease our live and not to have to handle the “MainDicomTags” mode that is a lot more complex to handle.
BR,
Alain
Hi Alain,
We have configured our PACS in “Full” mode using the cache and it’s works very well, Thanks for such a great help.
However, there are a subset of studies that still loads slow, this is because that studies have Overlay metadata, so, our viewer makes many requests to /dicom-web/studies/[StudyInstanceUID]/series/[SeriesInstanceUID]/instances/[SOPInstanceUID]/bulk/600X3000 endpoints.
Looking up at the code of the DicomWeb plugin we realized that every time bulk overlay data is required, the file is generated and then it’s sent to the client, but this is a slow operation and blocks the rendering of the study.
There is a way to generate a cache of this bulk data and store it in cache when the images are received just like it’s done with the dicoms tags metadata?
Thanks in advice,
Best regards
Hi Rodrigo,
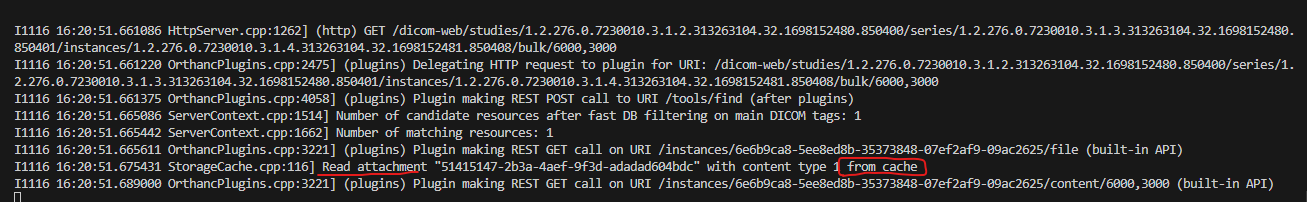
Here are the logs I have when calling
curl http://localhost:8043/dicom-web/studies/1.2.276.0.7230010.3.1.2.313263104.32.1698152480.850400/series/1.2.276.0.7230010.3.1.3.313263104.32.1698152480.850401/instances/1.2.276.0.7230010.3.1.4.313263104.32.1698152481.850408/bulk/6000,3000 --output /tmp/bulk
As you see, there is another cache involved here (the Orthanc StorageCache):
// Maximum size of the storage cache in MB. The storage cache
// is stored in RAM and contains a copy of recently accessed
// files (written or read). A value of "0" indicates the cache
// is disabled. (new in Orthanc 1.10.0)
"MaximumStorageCacheSize" : 128,
Can you verify your logs when calling this route ?
Do not hesitate to increase the cache size to 2000 (2 GB) or even more.
With the case enabled, Orthanc needs around 30 ms to answer my curl request…
HTH,
Alain.
Hi Alain,
Thanks for such a great response, we tested to enable the storage cache and effectively we get better response times. Unfortunately, we’ve use Orthanc on a Kubernetes cluster and horizontally replicated, therefore a cache stored in RAM is not suitable for us.
Despite that we finally achieve to solve this problem updating to the latest Ohif version, it load efficiently the Overlay data without blocking the study render.
Thanks for your help, I’ll to close this thread
Best regards.