Hello Everyone,
I have a folder of png images. Each image represents skin lesion.
I would like to convert these png images to dicom format.
Can you please let me know if there any converter available to transform this png to dicom image?
Thanks
Selva
Hi,
Many DICOM Viewers can convert .png file into DICOM file.
I developed HorliX, my customized version of Horos/OsiriX.
HorliX/Horos/OsiriX maybe can do it using plugin.
See attached files.
Then, you should transferred these dicom files into Orthanc, conquest,
dcm4chee...etc.
Ref: https://www.youtube.com/watch?v=83cSqXo_cLs
Hiroaki Inomata
MD: psychiatry
BSc: physics
PHAZOR, LLC
https://phazor.info
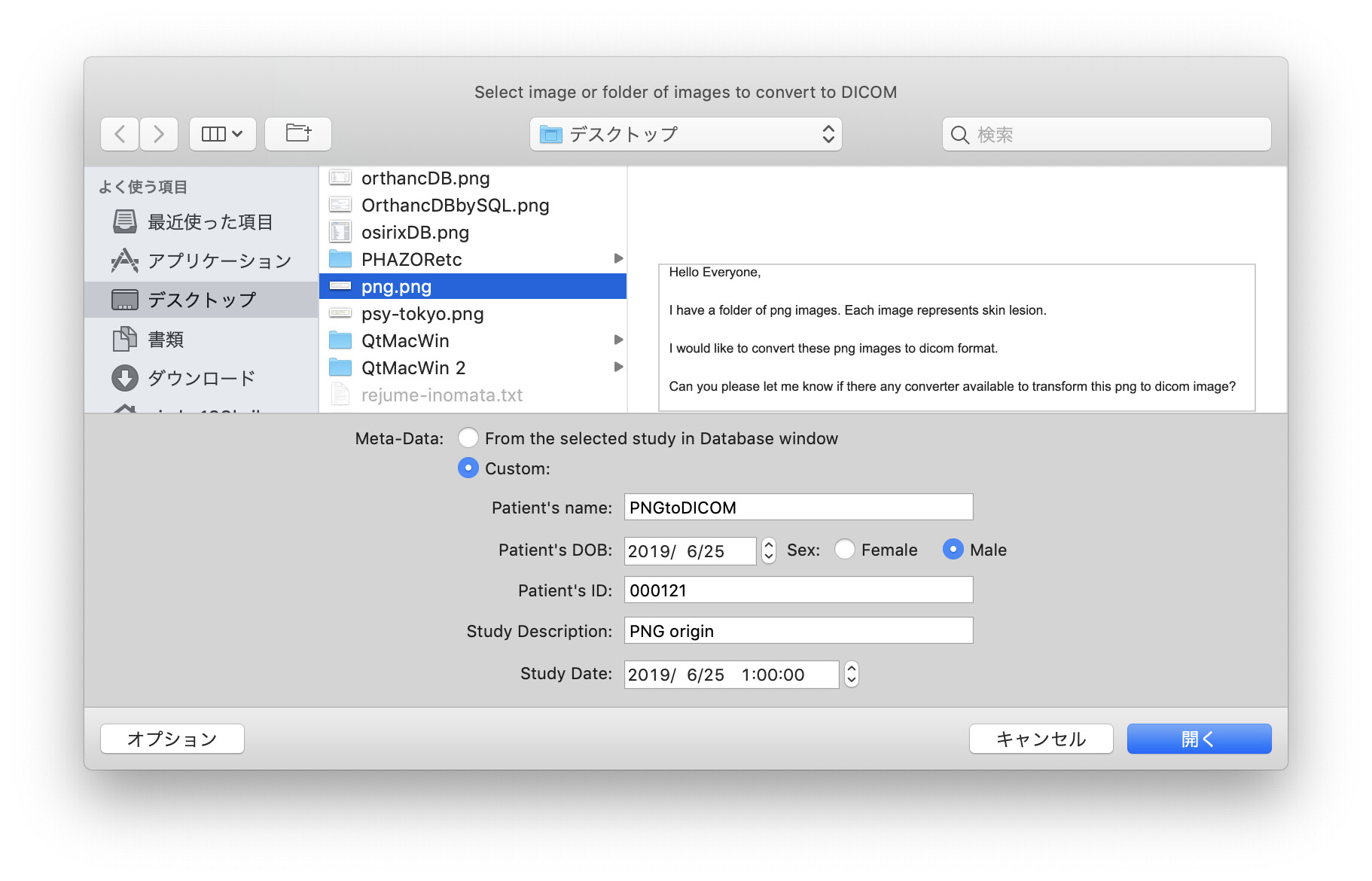
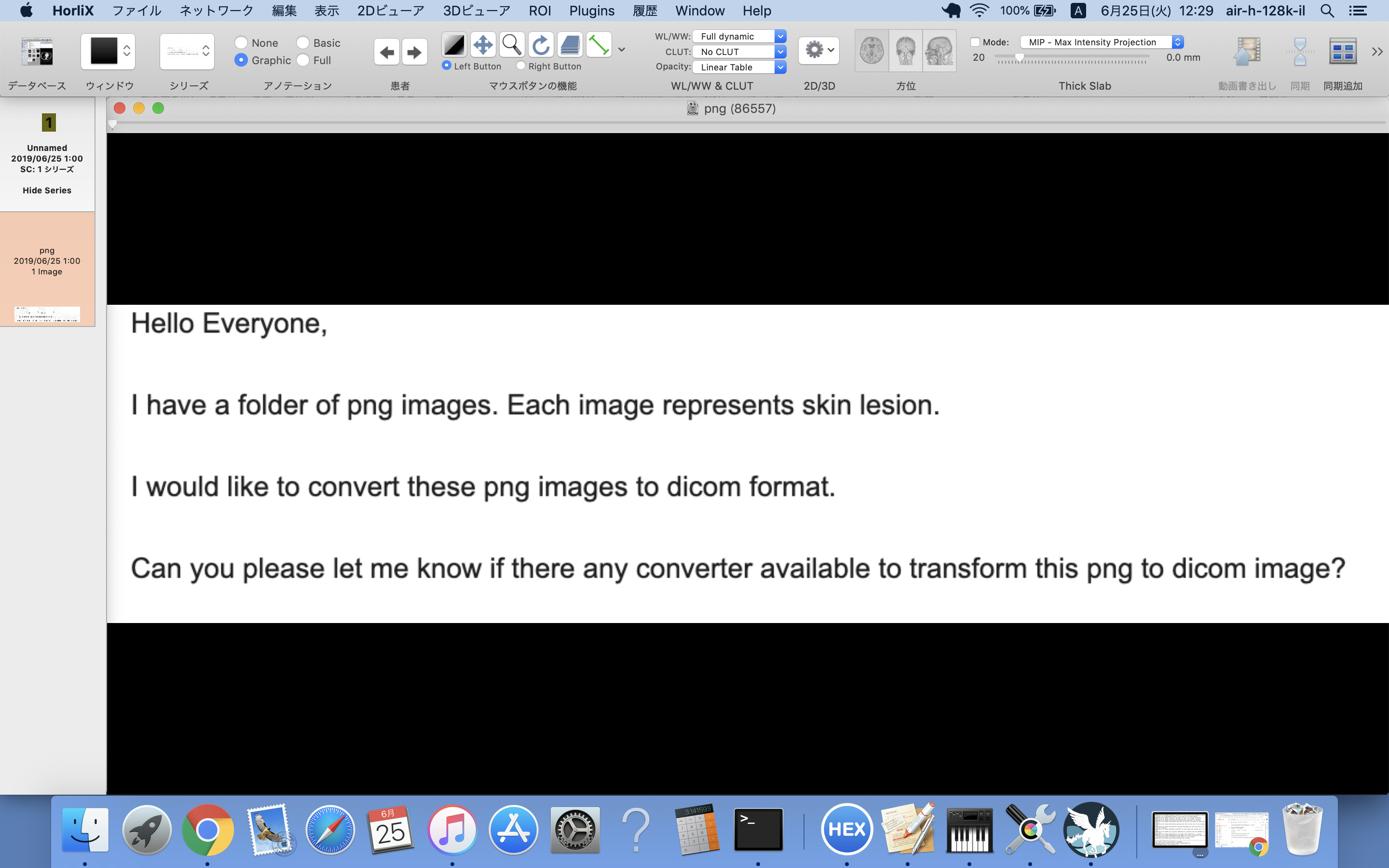
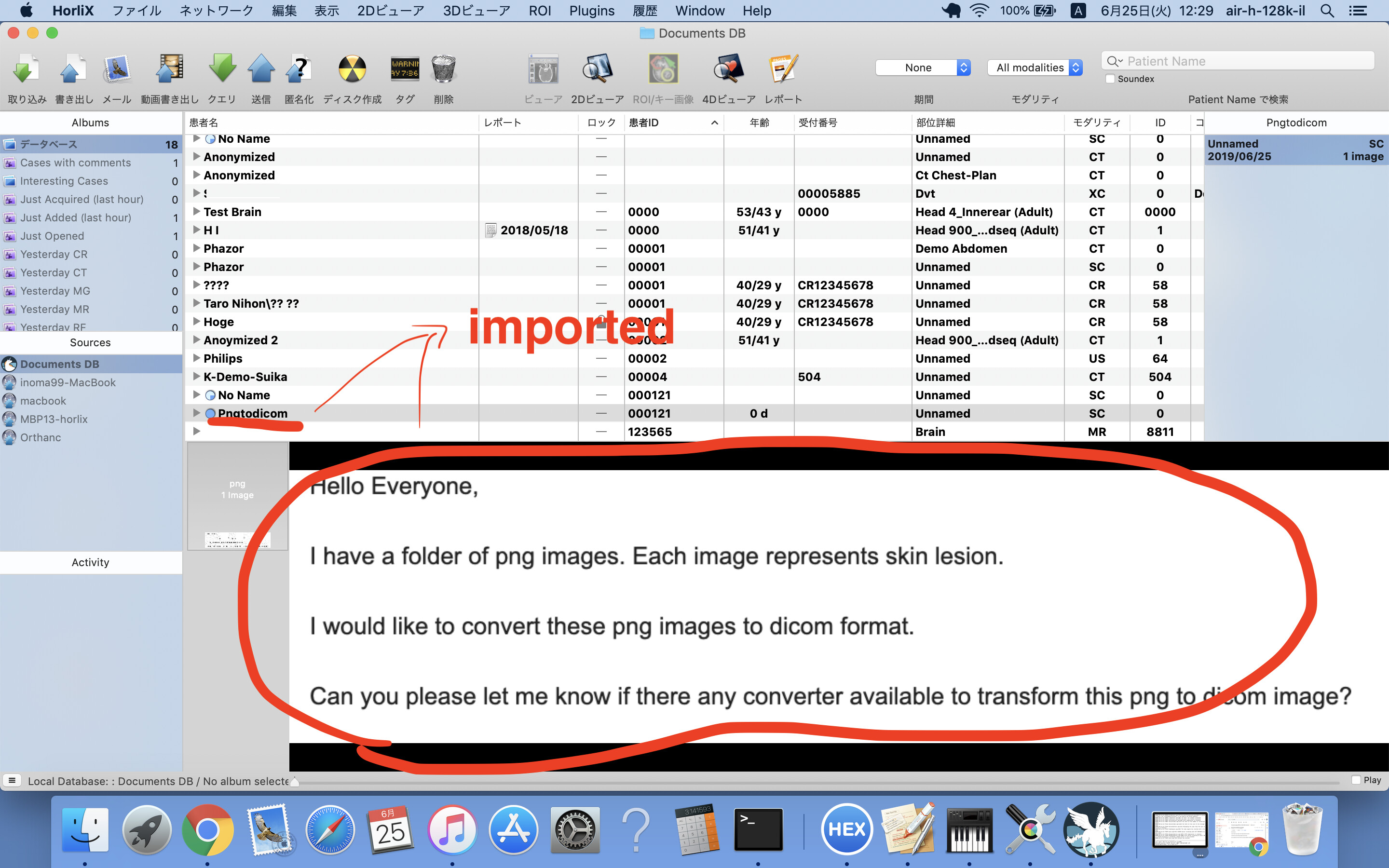
Hello Hiroaki Inomata,
Thank you for the help. Can I know whether the below two things is posible?
-
png to mhd format?
-
dicom to mhd format?
Is there any converter to do the same? I did search online but found something like SimpleITK which requires us to code
Thanks
Selva
Hi,
I am not familiar with mhd format.
Could you present mhd format and associated resources on internet?
Hiroaki Inomata
PHAZOR, LLC
https://phazor.info
Hi,
mhd format is ITK-specific one?
Ref: https://itk.org/Wiki/ITK/MetaIO/Documentation
In that page, I fount sample .mhd like:
2) dicom to mhd format?
I think that it can do in various ways.
DCMTK, well-known great free software, be able to extract DICOM tag
information from DICOM file.
You can get a number of column pixels, for example, in DICOM image
like the following code:
------------------ C++
Hello Hiroaki,
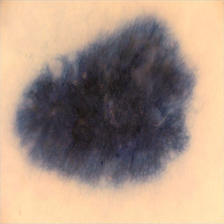
I have attached a skin lesion image which is in png format of 2 dimensions which is 224,224. But then for color, I guess it can be 224,224,3
When I convert this to dicom, it becomes 224,224,1. How can I retain it to be 224,224,3?
img.png - 224,224,3
img_1.dcm - 224,224,1
How can I make img_1.dcm as 224,224,3?
Can you please help?
img_1.dcm (148 KB)
I guess that (224, 224, 3) in mhd format is (columns, rows, a number of slices).
3 does not mean photmetricinterpretation(RGB).
Hello,
You can do this directly using the REST API of Orthanc, with URI “/tools/create-dicom”. Here is a full sample script in bash:
#!/bin/bash
set -ex
IMAGE=cat Lena.png | base64 -w 0
cat < /tmp/request.json
{
“Tags” : {
“PatientID” : “123”,
“PatientName” : “LENA”,
“StudyDescription” : “FROM^PNG”
},
“Content” : “data:image/png;base64,${IMAGE}”
}
EOF
curl -H ‘Expect:’ http://localhost:8042/tools/create-dicom -d @/tmp/request.json
Note that the integration tests of Orthanc contain more samples (notably to create a DICOM instance from a PDF file):
https://bitbucket.org/sjodogne/orthanc-tests/src/default/Tests/Tests.py
HTH,
Sébastien-
Hello Hiroaki,
Ahh. But can you let me know how can I generate 3 slices from one png image?
Thanks
Selva
Do you want to get a kind of DICOM files I attached?
Ref: skin224.zip
skin3DVR.png is 3D image based on the above DICOM files.
Hiraoki Inomata
PHAZOR, LLC
https://phazor.info
SKIN224.zip (978 KB)
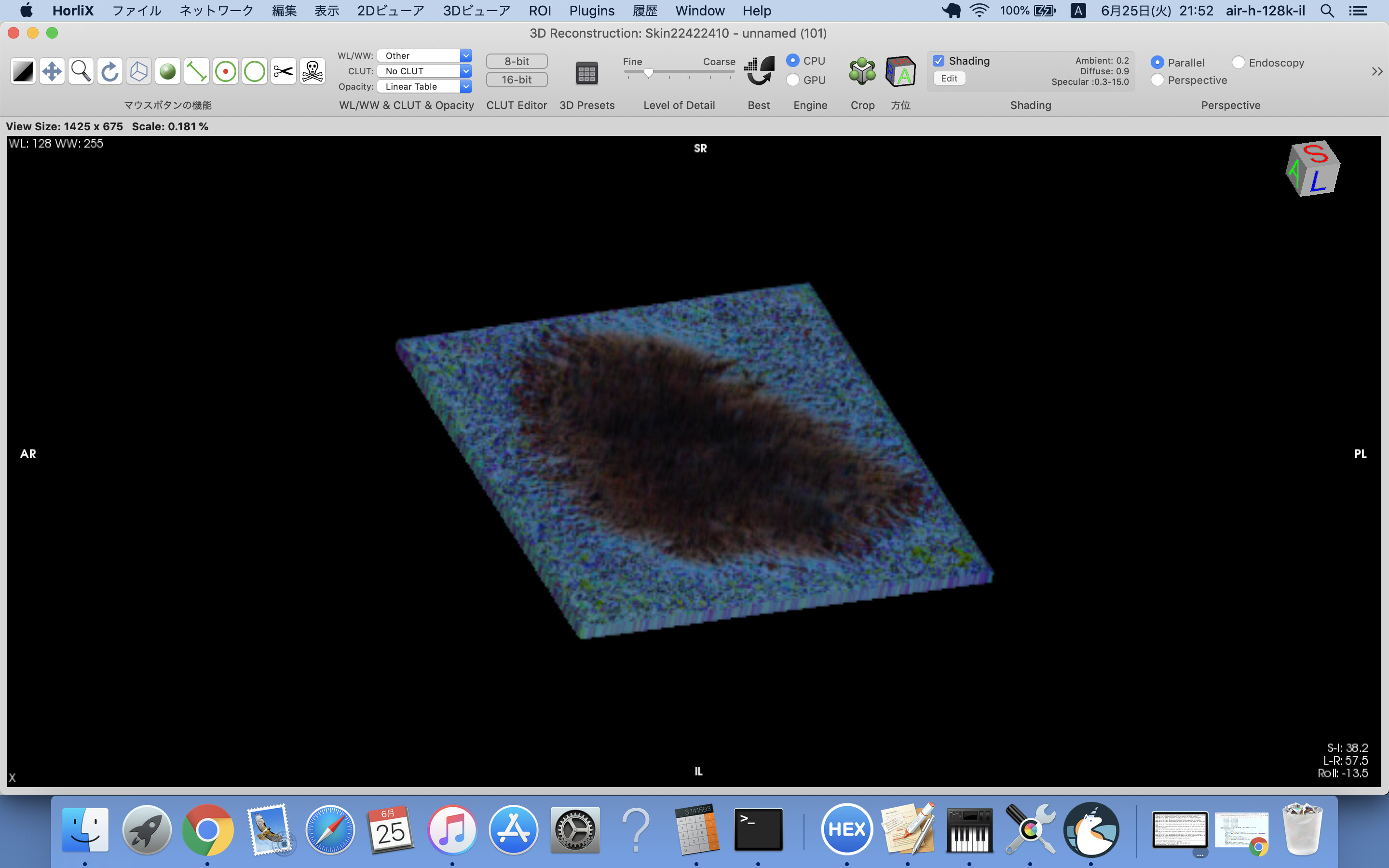
I mistook the RGB order of the pixel in the DICOM files of previous post.
Skinrgb.zip is, maybe, correct.
skinRGB-3DVR.png is 3D image constructed by the above DICOM files.
Hiroaki Inomata
MD: psychiatry
BSc: physics
PHAZOR, LLC
https://phazor.info
Skinrgb.zip (979 KB)
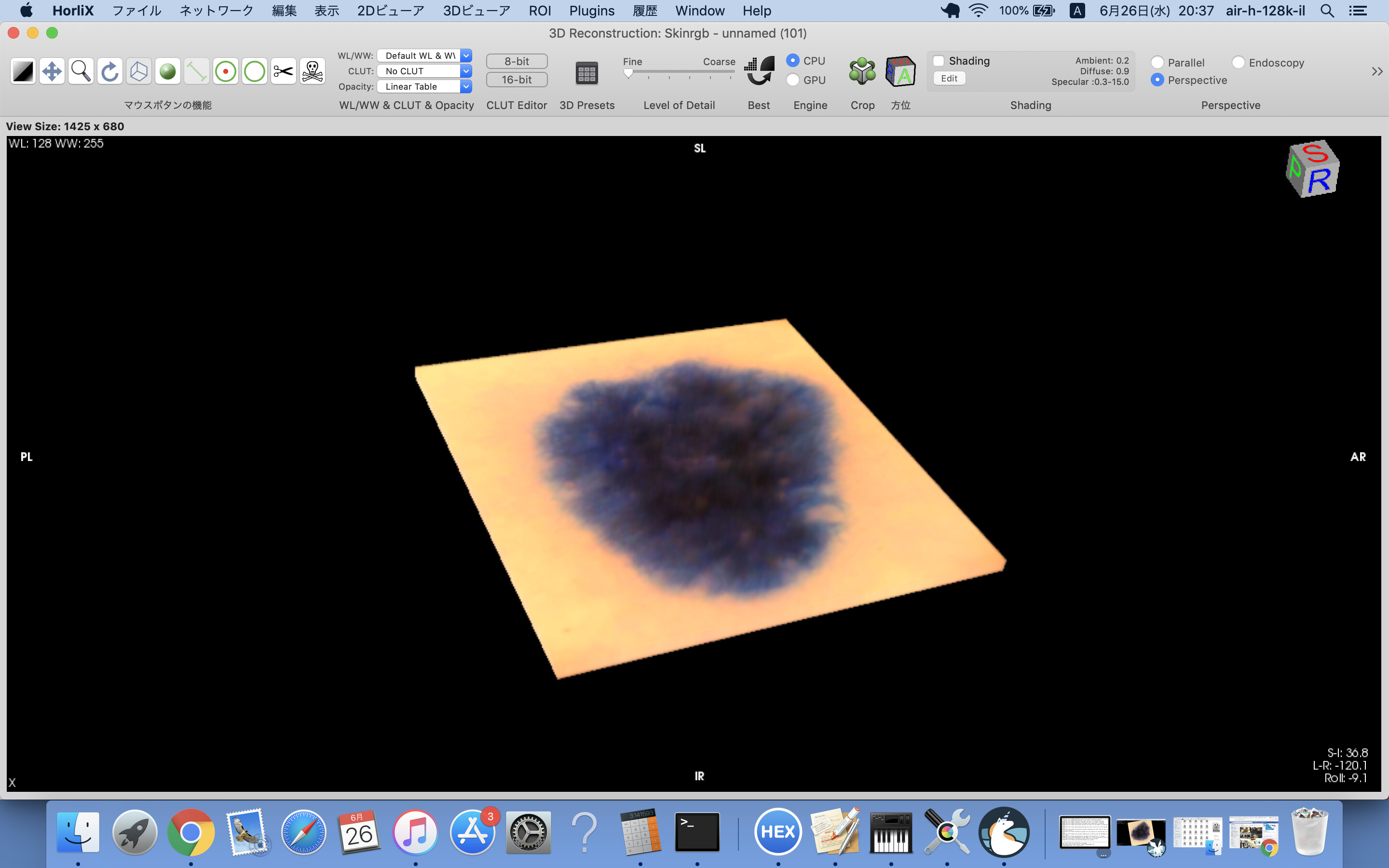
Hi,
BTW, what disease does this patient have?
I suspect just a mole or melanoma but I am not sure which is correct
because I am a just psychiatrist and not familiar with dermatology.
Hiroaki Inomata
MD: psychiatry
BSc: physics
PHAZOR, LLC
https://phazor.info





