Hello Everyone,
I am using Nvidia Clara deploy sdk to run the medical imaging workflow.
As a part of the workflow, from Orthanc, Clara picks up the dicom images, segments them and then sends the results back to Orthanc.
Though this works perfectly fine, “Orthanc web viewer” doesn’t display the segmented image output properly. I tried increasing and decreasing the contrast levels, zooming in and out but still I couldn’t view the segmented images output properly.
Can you please suggest me how to make this work?
IF you think I have to switch to any other viewer, can you please share any step-step guide on how to do this?
I am new and your inputs would definitely help
Thanks
Selva
Hi, Mr. Selva
You are dealing with nice work.
However, this is Orthanc Mailing List.
Do you know proper mailing list, forum or so in which we could discuss
more generalized Med x Tech problems like various Medical Open Source
Softwares, medical AI, Computer-Aided Diagnosis, medical image
processing, new devices...etc?
Hiroaki Inomata
MD: psychiatry
BSc: physics
PHAZOR, LLC
https://phazor.info
Please share a (possibly anonymized) sample DICOM file if you expect help from someone on this forum.
Hello Sébastien,
Here is the sample file attached that you requested
Thanks
Selva
IM0 (257 KB)
Hello,
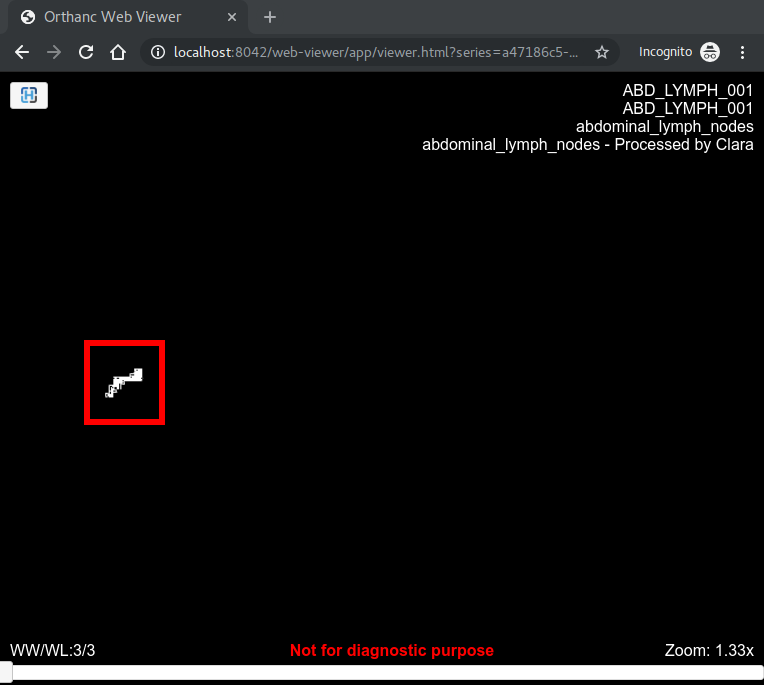
Your image only contains 2 values (0 and 5), as can be seen using this simple Python3 script:
import os
import pydicom
ds = pydicom.dcmread(‘IM0’)
values = {}
for n,val in enumerate(ds.pixel_array.flat):
if val in values:
values[val] += 1
else:
values[val] = 1
print(values)
The expected display is thus to see only white and black pixels. This is exactly what is shown by the Orthanc Web viewer, as shown in the attached screenshot (check out the red rectangle). So, again, there is nothing wrong here with Orthanc.
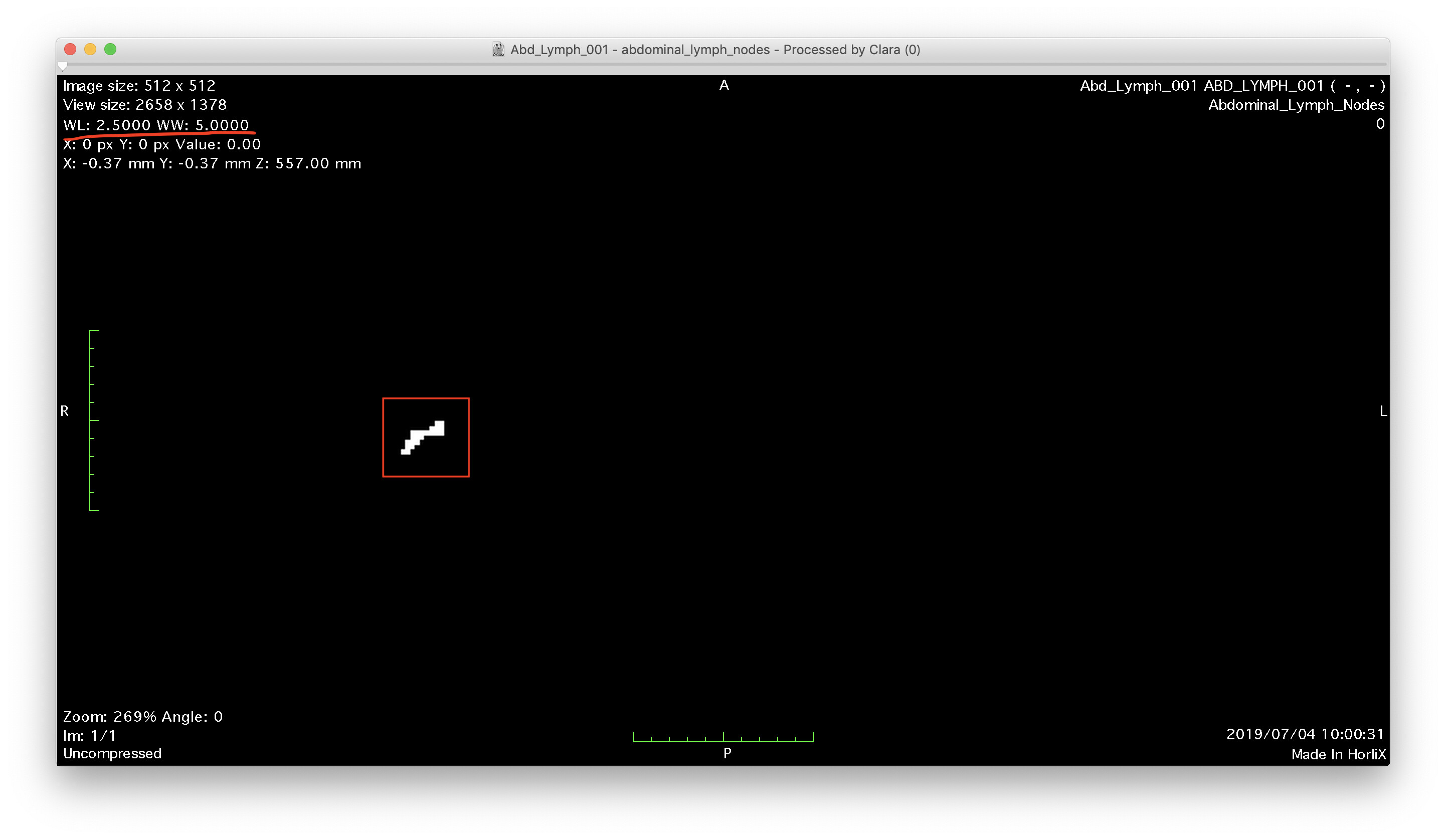
Hi,
It is one way Sebastian showed in the previous post.
Another way, for example, is that you should set WW=5.0 and WL=2.5
for your DICOM Viewer.
I guess that Selva want to extract lymph nodes and get binarization image of it
If so, setting the max value of the pixels in WW is another potent way.
I hope that the workflow would predict staging of certain disease precisely.
I am also interested in that kind of problem.
Hiroaki Inomata
MD, BSc
PHAZOR, LLC
https://phazor.info