Jakob,
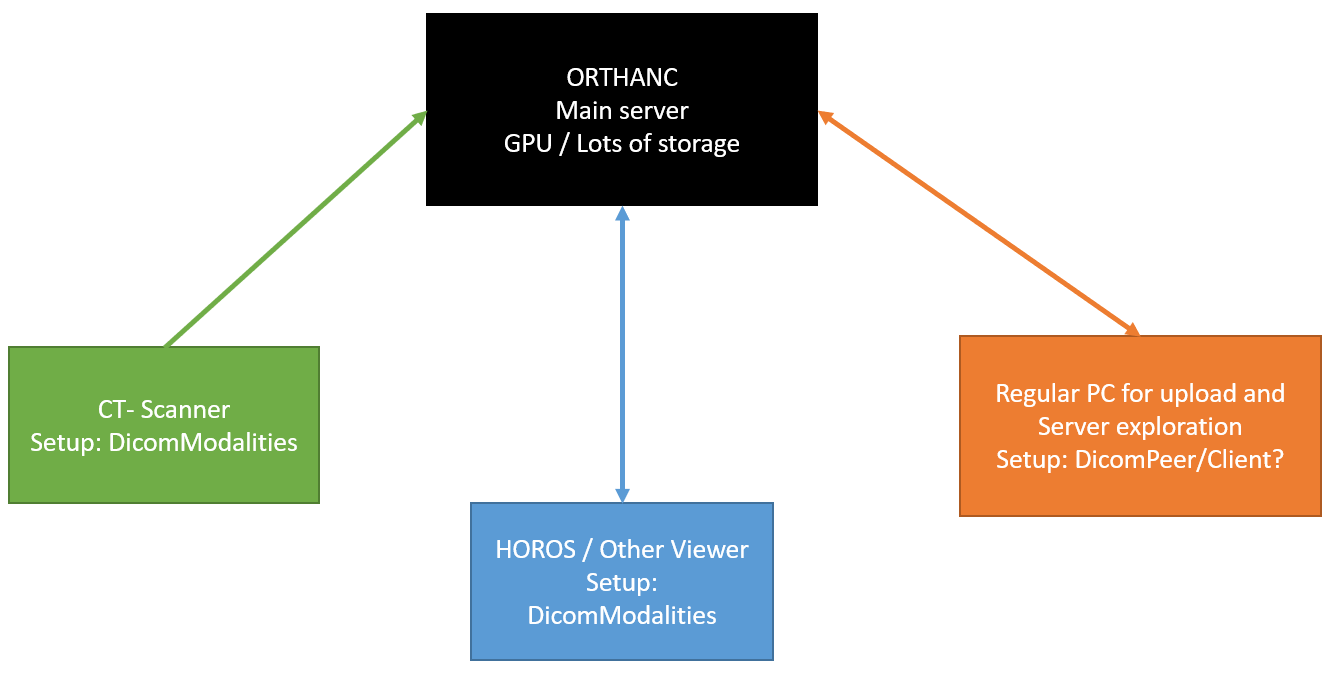
There are probably multiple ways to accomplish something like you are suggesting. It might depend a little upon where your users are located and what kind of access controls you need. It makes it a little easier if it is within the same institution and you don’t want fine access controls, and having a single instance of Orthanc running on decent hardware would probably be easier than maintaining multiple instances of Orthanc in a distributed network, although that can be done also. If the researches all have something like HOROS or OSIRIX, they can be networked with Orthanc pretty easily really. I think HOROS is sort of like a mini-PACS anyways. I have that also on Mac OSX and it works well with Orthanc.
I’ve actually been working on some ideas for something similar to that as part of a RIS project. I have a GitHub project for that, although I have to update the source code there.
A live demo is here: https://sias.dev:8000/browsestudies/index.html, which is a studies browser. I’ll have to write up some documentation, but it basically allows you to list all of the studies on the Orthanc Server in a paginated format, and you can search for studies, filtering on quite a few different parameters now. You can also view the studies in the Osimis Viewer or OHIF (this little magnifying glass to the left of study in the list. The little reports icon doesn’t do much here, but I do have a reporting feature on the RIS that I’ll try to move there. The litter airplane is under development, but would basically allow you to fetch a study and route it to a modality or AET, like a workstation. The “DCM” and “ZIP” download image archives. The “ALL” thing doesn’t do anything here, but I’ll make it show all of the studies for that particular patient. Patient History if the the RIS, but there is a DICOM field for “Additional Patient Info”, that I. might be able to display there if the RIS populates that. You’d have to play around with the Date Selector and other select lists at the top.
There is a much busier page here: https://sias.dev:8000/browsestudies/restapi.html
Basically a dev tool and API tester, although there is demo there about creating a PDF from HTML and then attaching that to a study.
(createPDF, Attach to Study, Open After Created). If you do that it’ll attach the PDF to the study, and when you view there will be a little document icon as an instance. If you drag that to the viewer it’ll display the PDF. You are welcome to try. It does that for just one study, so I check every now and then to delete any that were added.
It is an open site for now, so you are welcome to play with that.
Also, just generally, the demo is using the “ServeFolders” plug-in for that front-end, something that you or your developers might find useful because you can basically build your own front-end.
The demo is also running behind an NGINX or OpenResty server as a reverse proxy. There are some authentication tools for that kind of setup that can allow you to implement some sort of access controls, and you could actually provide access to the Orthanc Explorer through such a mechanism to remote users. I have that turned off now, and still exploring how best to do that (e.g. Oauth or something else). It isn’t that difficult to setup really, but the authentication part requires some thought. Osimis has an “Advanced Authentication” Plug-in that you might find useful.
/sds