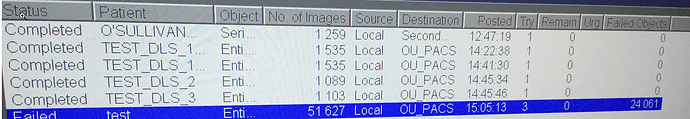
I have Orthanc receiving DICOMs from a MRI using a modified version of AutoClassify.py to write each instance out to disk.
For several small MRI’s (1600-3000 images) it works fine, however, when I do a test with a 51627 images it is failing:
Once this happens Orthanc seems to be automatically restarted every 14-20 minutes going from the logs (which contain no useful hints).
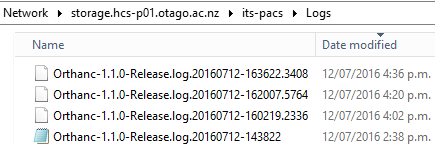
From AutoClassify I get the following message:
Writing new DICOM file: \storage.hcs-p01.otago.ac.nz\its-pacs\DICOMExport\Unknown\QA Phantom NiCl - test\DMHDS_PHANTOM\MR - ep2d_fid 55 mins\1.3.12.2.1107.5.2.19.46231.2016071209113992632029081.dcm
Unable to write instance 614b7d22-8e421c89-8c5d2453-91d77e05-bcbe361d to the disk
Traceback (most recent call last):
File “C:\admin\DICOM-export\DICOM-export.py”, line 161, in
‘limit’ : 4 # Retrieve at most 4 changes at once
File “C:\admin\DICOM-export\RestToolbox.py”, line 58, in DoGet
resp, content = h.request(uri + d, ‘GET’)
File “C:\Admin\Python\lib\site-packages\httplib2_init_.py”, line 1314, in request
(response, content) = self.request(conn, authority, uri, request_uri, method, body, headers, redirections, cachekey)
File "C:\Admin\Python\lib\site-packages\httplib2_init.py", line 1064, in _request
(response, content) = self.conn_request(conn, request_uri, method, body, headers)
File "C:\Admin\Python\lib\site-packages\httplib2_init.py", line 987, in _conn_request
conn.connect()
File “C:\Admin\Python\lib\http\client.py”, line 826, in connect
(self.host,self.port), self.timeout, self.source_address)
File “C:\Admin\Python\lib\socket.py”, line 711, in create_connection
raise err
File “C:\Admin\Python\lib\socket.py”, line 702, in create_connection
sock.connect(sa)
ConnectionRefusedError: [WinError 10061] No connection could be made because the target machine actively refused it
Does anyone have any advice on how to troubleshoot what might be going on? Increase/decrease limit/something else?
Cheers,
Mark