If you want to try that you could add this as a method in a Python Plug-in Script, but you need to:
-
Install wkhtmltopdf in your Orthanc container, using .deb packages via wget that have the ‘QT’ support also.
-
Install dcmtk in your Orthanc container, or figure out how to call DcmtkBridge from the Plug-in, to make the dsr2html executable available.
-
Tweak the script to fit your configuration.
-
Tweak where it saves /development/temp.dcm /development/temp.html" in the container in your docker-compose, or mkdir
I extracted it from my master so there might be some stuff missing.
DOCKERFILE
FROM osimis/orthanc:22.9.2
ENV DEBIAN_FRONTEND=noninteractive
ENV HTTP_BUNDLE_DEFAULTS=false
ENV AC_BUNDLE_DEFAULTS=false
RUN mkdir /python
# /python is bound to the host folder ./orthanc_python, but Orthanc needs to be restarted to see changes.
# /lua-scripts already exists in the container, and bound to lua in this folder. No need to restart to see changes, at least seems that way.
RUN apt-get update && apt-get --assume-yes install -y
wget
xz-utils
fontconfig
libfreetype6
libjpeg62-turbo
libpng16-16
libx11-6
libxcb1
libxext6
libxrender1
xfonts-75dpi
xfonts-100dpi
xfonts-scalable
xfonts-base
*dcmtk *
libpq-dev
added libpq-dev for import psycopg2
# Below is for the ARM M1 Architecture, library to support the PDF functions
RUN dpkg -i wkhtmltox_0.12.6-1.buster_arm64.deb
# Below is for the AMD Architecture, library to support the PDF functions
RUN wget https://github.com/wkhtmltopdf/packaging/releases/download/0.12.6-1/wkhtmltox_0.12.6-1.buster_amd64.deb
RUN dpkg -i wkhtmltox_0.12.6-1.buster_amd64.deb
RUN pip3 install pydicom pynetdicom pdfkit imgkit hl7 wkhtmltopdf mysql-connector-python requests
RUN pip3 install psycopg2
COPY docker-entrypoint.sh /
python.py (Plug-in Script)
import shutil
import pdfkit # https://pypi.org/project/pdfkit/, sudo python3 -m pip install pdfkit
from pdfkit import configuration
from pdfkit import from_string
import pydicom # https://github.com/pydicom/pydicom, sudo python3 -m pip install pydicom
from pydicom.datadict import dictionary_keyword
from pydicom import dcmread, dcmwrite
from pydicom.filebase import DicomFileLike
from pydicom.dataset import Dataset, FileDataset, FileMetaDataset
from pydicom.uid import ExplicitVRLittleEndian, generate_uid
def ReceivedInstanceCallback(receivedDicom, origin):
Only do the modifications if via DICOM and ideally filter by AET.
orthanc.LogWarning('DICOM instance received in ReceivedInstanceCallback from ’ + str(origin))
dataset = dcmread(BytesIO(receivedDicom))
jsonTags = json.loads(orthanc.DicomBufferToJson(receivedDicom, orthanc.DicomToJsonFormat.HUMAN, orthanc.DicomToJsonFlags.NONE, 0))
orthanc.LogWarning(json.dumps(jsonTags, indent = 2, sort_keys = True))
if origin == orthanc.InstanceOrigin.DICOM_PROTOCOL:
Do Nothing for now
return orthanc.ReceivedInstanceAction.KEEP_AS_IS, None
elif origin == orthanc.InstanceOrigin.REST_API:
if “Modality” in jsonTags and jsonTags[‘Modality’] == “SR”:
orthanc.LogWarning("NEW SR INSTANCE VIA RESTAPI: "+json.dumps(jsonTags, indent = 2, sort_keys = True))
logging.info(“NEW SR INSTANCE VIA RESTAPI”+json.dumps(jsonTags, indent = 2, sort_keys = True))
If it an SR Modality type, use dcmtk dsr2html to convert to HTML, then
use wkhtmltopdf to convert to an encapsulated PDF for easier display.
pathtobinary = shutil.which(“dsr2html”)
dataset.save_as(“/development/temp.dcm” ,write_like_original=True)
cmd = pathtobinary + " -Ei /development/temp.dcm /development/temp.html"
os.system(cmd) # returns the exit status
pathtobinary = shutil.which(“wkhtmltopdf”)
config = pdfkit.configuration(wkhtmltopdf=pathtobinary)
options = {
‘page-size’: ‘Letter’,
‘margin-top’: ‘0.75in’,
‘margin-right’: ‘0.75in’,
‘margin-bottom’: ‘0.75in’,
‘margin-left’: ‘0.75in’,
‘footer-line’:‘’,
‘footer-font-size’:‘12’,
‘footer-center’: ‘Page [page] of [toPage], [date]’,
‘encoding’: ‘utf-8’
}
pdf = pdfkit.from_file**(“/development/temp.html”**, False,options=options)
encoded = base64.b64encode(pdf).decode()
dicomdata = dict()
dicomdata[‘Force’] = True
dicomdata[‘Tags’] = {
“PatientID”:jsonTags[‘PatientID’],
“PatientName”:jsonTags[‘PatientName’],
“PatientBirthDate”:jsonTags[‘PatientBirthDate’],
“PatientSex”:jsonTags[‘PatientSex’],
“Modality”:“OT”,
“SeriesDescription”: jsonTags[‘SeriesDescription’]+“, SR Converted to PDF”,
“SequenceName” : “NA”,
“ImageComments”:“SR Converted using dsr2html & wkhtmltopdf”,
“SOPClassUID”:“1.2.840.10008.5.1.4.1.1.7.4”,
“StudyInstanceUID”:jsonTags[‘StudyInstanceUID’]
}
dicomdata[‘Content’] = “data:application/pdf;base64,”+encoded;
convertedSR = json.loads(orthanc.RestApiPost(‘/tools/create-dicom’, json.dumps(dicomdata)))
return orthanc.ReceivedInstanceAction.MODIFY, dataset_to_bytes(dataset)
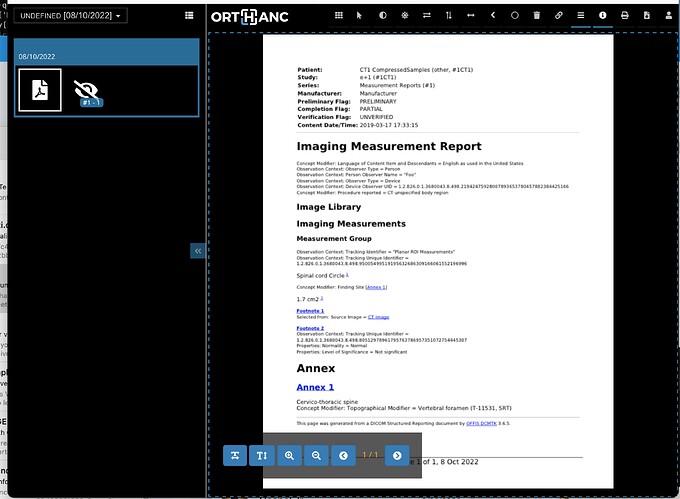
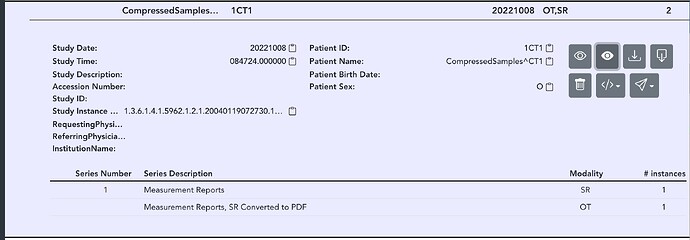
Load up a sample SR instance through the RESTAPI / Explorer
It should create a PDF version of the SR.
/sds