Hello Orthanc Community,
I am attempting to implement the Orthanc PACS in my biomedical research group. We are using the indexer plugin to load images directly from a directory, all within Docker containers. Additionally, we are connecting the OHIF viewer (alongside Orthanc’s own plugin, as we have developed new modes and extensions that we connect to the viewer) and a Keycloak layer.
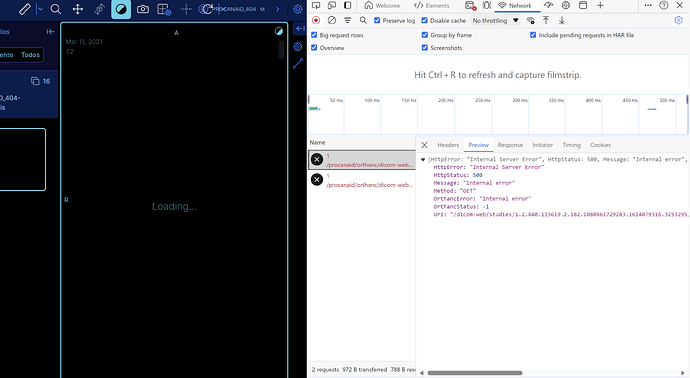
The scanning functionality works normally, and we have our entire database organized and loaded in Orthanc, viewable in the UI Explorer 2. The problem arises when we use the versioned Docker images from orthancteam/orthanc or jodogne/orthanc-plugins with specific version numbers (e.g orthancteam/orthanc:24.5.1, and not the latest from jodogne, which is in the mainline). The images with versioned numbers do not display in the OHIF viewer, and the verbose logs only show the following:
[HTTP-27 PluginsErrorDictionary.cpp:100] Exception inside the plugin engine: Internal error
Conversely, when we use the latest version from jodogne and mainline for all plugins, the images display in the OHIF viewer but with significant delay. This is because, despite enabling accept transfer-syntax=* and configuring OHIF to accept all images, Orthanc in the mainline version decides to transcode the image. This results in Orthanc becoming busy and unresponsive to DICOMweb when dealing with studies containing many instances.
Logs from orthancteam/orthanc:24.5.1:
2024-05-24 19:01:07 I0524 17:01:07.280106 HTTP-48 OrthancPlugins.cpp:2475] (plugins) Delegating HTTP request to plugin for URI: /dicom-web/studies/1.2.840.113619.2.182.1080861729283.1614079316.3253295/series/1.2.840.113619.2.312.4120.8398801.12801.1615358812.516/instances/1.2.840.113619.2.312.4120.8398801.12113.1615358911.743/frames/1
2024-05-24 19:01:07 I0524 17:01:07.280217 HTTP-48 OrthancPlugins.cpp:4058] (plugins) Plugin making REST POST call to URI /tools/find (after plugins)
2024-05-24 19:01:07 I0524 17:01:07.281616 HTTP-48 ServerContext.cpp:1561] Number of candidate resources after fast DB filtering on main DICOM tags: 1
2024-05-24 19:01:07 I0524 17:01:07.281774 HTTP-48 ServerContext.cpp:1709] Number of matching resources: 1
2024-05-24 19:01:07 I0524 17:01:07.281862 HTTP-48 PluginsManager.cpp:161] (plugins) DICOMweb RetrieveFrames on b87d9910-1a42a153-dd95fa37-348cd849-6cf4f282, frames: 1
2024-05-24 19:01:07 I0524 17:01:07.281882 HTTP-48 OrthancPlugins.cpp:3221] (plugins) Plugin making REST GET call on URI /instances/b87d9910-1a42a153-dd95fa37-348cd849-6cf4f282/metadata/TransferSyntax (built-in API)
2024-05-24 19:01:07 I0524 17:01:07.282018 HTTP-48 OrthancPlugins.cpp:3221] (plugins) Plugin making REST GET call on URI /instances/b87d9910-1a42a153-dd95fa37-348cd849-6cf4f282/file (built-in API)
2024-05-24 19:01:07 I0524 17:01:07.282178 HTTP-48 StorageCache.cpp:127] Read attachment "43a6212c-ed49-4094-aa71-93b05f057713" with content type 1 from cache
2024-05-24 19:01:07 E0524 17:01:07.282483 HTTP-48 PluginsErrorDictionary.cpp:100] Exception inside the plugin engine: Internal error
Logs from jodogne/orthanc-plugins:latest:
2024-05-24 19:12:41 I0524 17:12:41.173695 HTTP-0 OrthancPlugins.cpp:3223] (plugins) Plugin making REST GET call on URI /instances/370ee877-c6115228-6ae6c80e-8b174c56-cfb53c04/metadata/TransferSyntax (built-in API)
2024-05-24 19:12:41 I0524 17:12:41.173710 HTTP-49 ServerContext.cpp:1713] Number of matching resources: 1
2024-05-24 19:12:41 I0524 17:12:41.173744 HTTP-0 dicom-web:/WadoRsRetrieveFrames.cpp:483] The file is in a transfer syntax 1.2.840.10008.1.2.2 that is not allowed by the DICOMWeb standard -> it will be transcoded to Little Endian Explicit
2024-05-24 19:12:41 I0524 17:12:41.173766 HTTP-0 OrthancPlugins.cpp:3223] (plugins) Plugin making REST GET call on URI /instances/370ee877-c6115228-6ae6c80e-8b174c56-cfb53c04/file?transcode=1.2.840.10008.1.2.1 (built-in API)
2024-05-24 19:12:41 I0524 17:12:41.173771 HTTP-49 dicom-web:/WadoRsRetrieveFrames.cpp:461] DICOMweb RetrieveFrames on da9d67d5-d40f5d13-1f0bc073-c04076d1-04e6a619, frames: 1
2024-05-24 19:12:41 I0524 17:12:41.173779 HTTP-49 OrthancPlugins.cpp:3223] (plugins) Plugin making REST GET call on URI /instances/da9d67d5-d40f5d13-1f0bc073-c04076d1-04e6a619/metadata/TransferSyntax (built-in API)
2024-05-24 19:12:41 I0524 17:12:41.173861 HTTP-1 dicom-web:/WadoRsRetrieveFrames.cpp:483] The file is in a transfer syntax 1.2.840.10008.1.2.2 that is not allowed by the DICOMWeb standard -> it will be transcoded to Little Endian Explicit
2024-05-24 19:12:41 I0524 17:12:41.173805 HTTP-3 dicom-web:/WadoRsRetrieveFrames.cpp:483] The file is in a transfer syntax 1.2.840.10008.1.2.2 that is not allowed by the DICOMWeb standard -> it will be transcoded to Little Endian Explicit
2024-05-24 19:12:41 I0524 17:12:41.173897 HTTP-1 OrthancPlugins.cpp:3223] (plugins) Plugin making REST GET call on URI /instances/701695c5-89219984-e624813a-0c9ff702-9006e23f/file?transcode=1.2.840.10008.1.2.1 (built-in API)
2024-05-24 19:12:41 I0524 17:12:41.173904 HTTP-3 OrthancPlugins.cpp:3223] (plugins) Plugin making REST GET call on URI /instances/1bcc3a0a-8f52350f-5cf5d6f1-cfc813ad-b7dd0e94/file?transcode=1.2.840.10008.1.2.1 (built-in API)
2024-05-24 19:12:41 I0524 17:12:41.173956 HTTP-49 dicom-web:/WadoRsRetrieveFrames.cpp:483] The file is in a transfer syntax 1.2.840.10008.1.2.2 that is not allowed by the DICOMWeb standard -> it will be transcoded to Little Endian Explicit
2024-05-24 19:12:41 I0524 17:12:41.173996 HTTP-49 OrthancPlugins.cpp:3223] (plugins) Plugin making REST GET call on URI /instances/da9d67d5-d40f5d13-1f0bc073-c04076d1-04e6a619/file?transcode=1.2.840.10008.1.2.1 (built-in API)
Screenshots:
We also tested in OHIF Viewer and everything is very quick and smooth without the issues we encountered when connecting to the DICOMweb of Orthanc.
Summary
In summary, dicom images from our center, whether anonymized or not and exported correctly from the MR machine, exhibit this issue. Ideally, we would like the images to display in the OHIF viewer without transcoding.
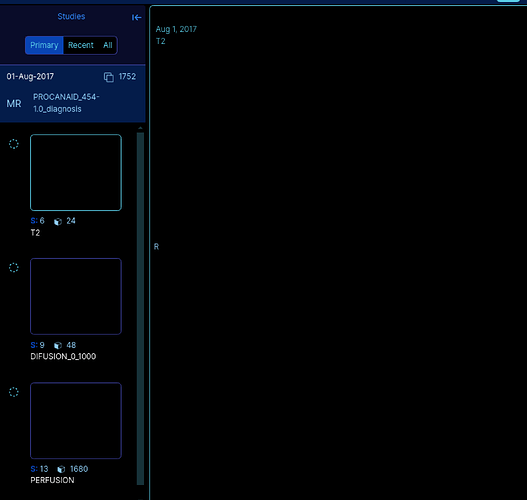
We are providing an anonymized image; it is a T2 of prostate scan: Example
Thank you for your assistance and your great job!!