Hello Orthanc Team,
We are facing an issue with our OHIF-Orthanc-PostgreSQL setup. We have successfully configured Orthanc with the PostgreSQL plugin, using it only as an indexer, and set up Orthanc to scan a folder with the Indexer plugin. Initially, everything worked as expected. Additionally, we connected Orthanc to OHIF using a reverse proxy through Nginx, enabling HTTPS for the communication between OHIF and Orthanc. All connections were stable, and the system was running smoothly.
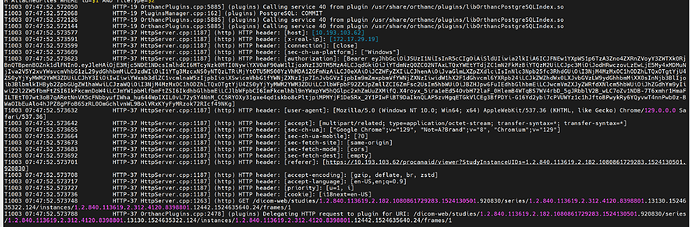
However, after an indeterminate period of time, the OHIF viewer stops functioning, and requests at the series/instance level to Orthanc start failing as (pending to finally timeout). We suspect the issue lies in the connection between Orthanc and PostgreSQL, but despite enabling all available logs, we haven’t been able to identify the root cause. This is the only error we found.
And trying to open a study, with logs of postgres we only see this:
For context, we are working with a large dataset of over 4,000 studies and approximately 1TB of data. Our setup was deployed using Docker Compose with PostgreSQL 16.4 and the latest version of Orthanc.
Initially, we thought the problem was due to an overload of queries to the database, but after conducting load tests and successfully opening numerous studies, the system continued to perform well. The issue consistently arises after an undetermined period, where it seems the connection between Orthanc and PostgreSQL is being closed unexpectedly.
Below is the relevant part of our Orthanc configuration file:
procanaid-orthanc.json (59.6 KB)
Any insights or recommendations on how to address this issue would be greatly appreciated.
Best regards,
Adrián
I was able to catch the error thanks to the trace logs, and now I don’t believe the issue is with PostgreSQL (sorry for the earlier assumption).
When ohif-orthanc is functioning properly, I see logs like this:
However, when it stops working, I see logs like this:
Here is my current nginx configuration related to Orthanc and OHIF:
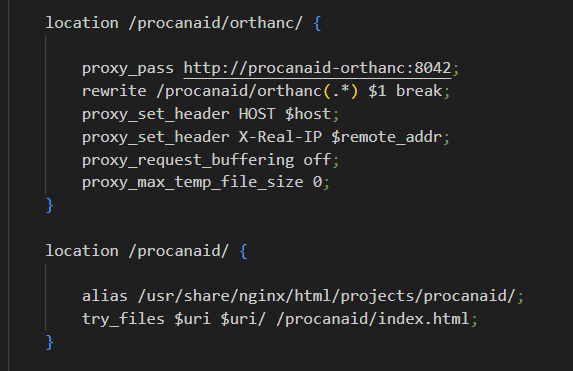
To add some context, nginx is running outside the Orthanc container. Could this issue be related to something I need to configure in nginx, or is it something within the Orthanc container?
Additionally, I have a Keycloak setup, but it only affects Orthanc Explorer 2. The API is not yet secured—neither with orthanc-auth-service nor with OAuth2, which is something I plan to implement later.
Thank you very much for your support!
Hi Adriàn,
Difficult to analyze the logs from such small extract and actually it’s not clear what you mean by “when OHIF stops working”.
So all I can share right now is the standard config we use in nginx to reverse proxy Orthanc:
location /orthanc/ {
proxy_pass http://orthanc:8042;
rewrite /orthanc(.*) $1 break;
proxy_set_header Host $http_host;
proxy_set_header X-Forwarded-For $proxy_add_x_forwarded_for;
proxy_request_buffering off;
proxy_max_temp_file_size 0;
client_max_body_size 0;
}
client_max_body_size defaults to 8 MB which can be a source of problems when dealing with large instances.
HTH,
Alain
Thank you, @alainmazy.
We’ve implemented client_max_body_size at the server level as it applies to all our Orthanc locations. However, we’ll follow your suggestion and move it to the location block, adopting your nginx configuration.
To clarify my previous explanation, I’ve recorded a video demonstrating the issue. The video shows studies being opened successfully in OHIF and Orthanc. Everything works fine until the 7:45 mark, when it suddenly stops working. I then showcase the logs and Orthanc explorer, which display an error at the instance level. Finally, I attempt to demonstrate the same error using the API. Apologies for the video length, but it comprehensively captures the problem’s progression.
Orthanc-OHIF Demo. Unexpected Termination During Operation.mp4
Additionally, I’ve created a short video showing that restarting the Orthanc container resolves the issue and restores functionality.
Restoring Orthanc Functionality with Container Restart.mp4
Thank you so much for your help.
Best regards,
Adrián
Hello again,
I’ve continued researching this issue and may have found a related scenario. I can reproduce the same problem by disabling the indexer mode in the Orthanc configuration:
"Indexer" : {
"Enable" : true,
"Folders" : [ "/project/ADQUISICIONES" ], // List of folders to synchronize
"Interval" : 10 // Delay between two synchronizations
},
When disabled, files and tags at the instance level are not available. Is it possible that with large datasets indexed by the indexer plugin, random “disconnections” occur, causing the issue I’m experiencing (when it is enabled)?
Does the indexer plugin extra logs that I can enable?
Thank you for your assistance.
Hi there,
I have created two logs: one for when Orthanc works and I can explore the tags at the instance level in Orthanc Explorer 2, and another for when it doesn’t work.
When it works:
working.txt (23.6 KB)
When it doesn’t work:
not-working.txt (29.2 KB)
The only difference between the two logs is this line:
I1008 12:57:46.020260 HTTP-4 StorageCache.cpp:182] Read start of attachment "1182e40d-4a2f-4143-9a19-ed9ef79c14c9" with content type 1 from cache
I restart the Orthanc container and open a study that I haven’t opened before (trying to avoid using cached data), but when it works, it always says “content type 1 from cache”.
Thank you again for your help!
Well, first of all, I’m not a specialist of the orthanc-index plugin and I actually don’t see any errors in the ‘not-working.txt’ log.
You might not be aware that the orthanc-index plugin actually has its own SQLite database so when you try to access /instances/../tags, Orthanc will actually get the attachment UUID from PostgreSQL and then the path to the real file from SQLite.
So, when you disable the orthanc-index plugin, you won’t be able to access files anymore.
And, if you have multiple Orthanc instances connected to the same PostgreSQL, they won’t be able to connect to a unique orthanc-index SQLite DB → it might look like a DB disconnection if that happens.
HTH,
Alain
Thank you @alainmazy for your detailed explanation. I appreciate your insights.
Yes, I have multiple Orthanc instances connected to a single PostgreSQL container, but with separate databases for each Orthanc instance. Each instance, with its indexer plugin, scans a different project folder (one for prostate, another for lung, another for brain, etc.). In other words, each is linked to a different folder, so the Orthanc instances don’t interact with each other at any level.
From what you’re saying, I understand that with a large database, the indexer plugin works well for scanning the entire folder, but it’s limited by the SQLite database becoming very large. I gather there’s no way to increase resources to make the SQLite function better.
Given the nature of this error, is there anything I can do? The only option I see is to give up my filesystem folder organization that I was managing with the indexer plugin, and “re-upload” all the DICOMs directly to the PostgreSQL database. But is no ideal for me 
Is this understanding correct? Are there any other alternatives or solutions you might suggest?
Thank you again
No, right now, I have no alternative solutions.
In a few months, we shall be able to re-write the index plugin such that it does not use a separate SQLite DB.
Alain.
Thank you @alainmazy!
We managed to upload all DICOM files using ImportDicomFiles.py, and now both Orthanc and OHIF are working properly.
I was looking into the StoreDicom flag to run Orthanc in index-only mode, hoping that if I uploaded files within the same container as Orthanc, it would also save the path to the original DICOM files. However, this isn’t working as expected - I cannot download the DICOM instances:
I also noticed that running ImportDicomFiles twice with StoreDicom set to true duplicates the series within a study (which is undesirable).
Are there other ways to upload all DICOM files and monitor new files in a folder? Or is the indexer-plugin the only option?
If not, I’m considering creating a Python script that checks Orthanc before uploading any files.
Thanks you again, and I will waiting this re-write of the plugin 
This should not happen as long as the DICOM IDs inside the file are kept untouched between successive uploads. Re-uploading the same file is a very common use case and it should really not create duplicates.
I would strongly recommend you to restart a new setup without the indexer plugin and upload the file with “StoreDicom” kept to true. You can then upload folders with the python script or drop them in OE2.
Best regards,
Alain.
Hi @alainmazy,
Do you have any updates on the index plugin regarding support for PostgreSQL instead of SQLite?
Thanks a lot! 
Hi @adrian_galiana
No, sorry. The first version of the advanced-storage plugin is not yet ready and, I must admit that adding an indexer mode is not in our top priority list.
Best,
Alain
Thanks a lot, @alainmazy!
Even though the switch from SQLite to Postgres hasn’t been implemented yet, the indexer seems to be working better for me now.
Thanks for your hard work!