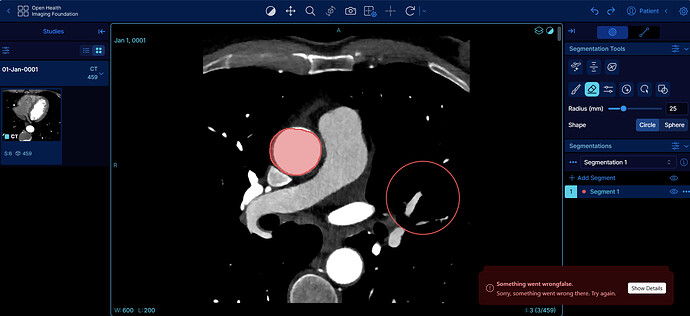
Hi everybody,
I’m having a problem during the export a modified DICOM SEG within the OHIF Viewer with segmentation tools. I thought that when I click this feature, the segmentation would have saved back to my Orthanc server, but OHIF shows a popup with the text “Something went wrong”. To face that I’m forced to download the DICOM SEG and upload to Orthanc. Why? Is that a problem relalted to some configuration in Orthanc or is that an OHIF issue?
Tks a lot,
Lorenzo
I too run into this issue. Clicking on the “Show Details” button simply dismisses the alert without providing any information. I don’t see any relevant lines in the logs. Not sure how to troubleshoot this, or whether this is an OHIF or Orthanc problem. Currently using an Orthancteam docker image, but also present with the latest orthanc-python image.
Hi,
it looks like a bug of OHIF Viewer ( [OHI-1445] fix(store-segmentation): storing segmentations was hitting the wrong command resulting in an undefined datasource by IbrahimCSAE · Pull Request #4755 · OHIF/Viewers · GitHub) I hope that soon wiill be released the new version and in Orthanc the updated plugin.
Lorenzo
Hi everybody, I am using the last orthanc image (25.5.1) that should use the OHIF 3.10.1 but I have still this problem during export of Dicom Seg
Can anybody give me any suggestion on why it gives this error?
Tks a lot,
Lorenzo
Hello,
Shouldn’t this question be asked to the OHIF team? The Orthanc project does not develop OHIF.
Regards,
Sébastien-