Hello,
If I issue the following command over your log file:
$ grep -i ‘Reading attachment’ Orthanc.log | sort | wc -l
20
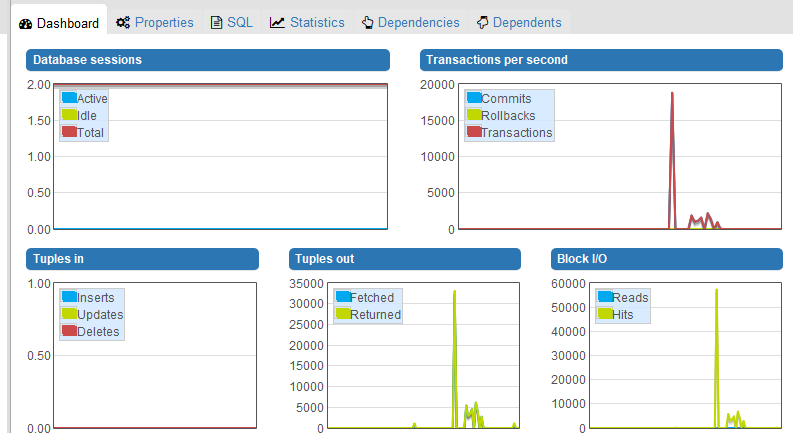
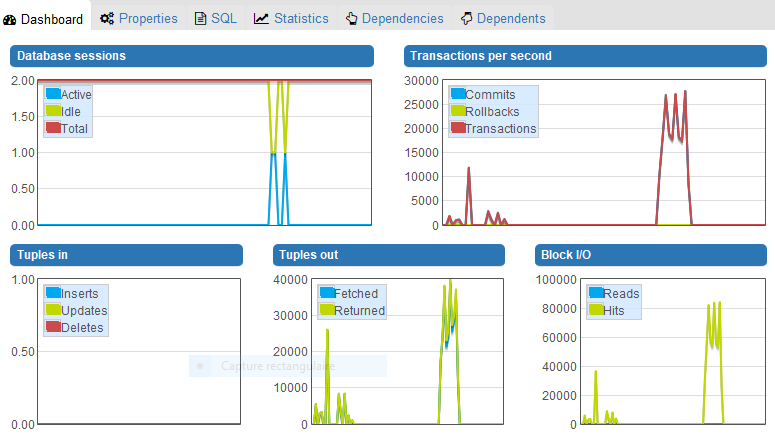
I observe that Orthanc only reads 20 files from the its database to fulfill the QIDO-RS request. All these files are JSON summaries, which means that no DICOM parsing is involved. Reading a number of JSON summaries that is twice the number of studies is the expected behavior: The QIDO-RS call first selects one instance for each study, then filters the tags of the selected instances from their JSON summary (which implies one file reading for each study), and finally creates an answer (which implies a second file reading for each filtered study).
As can be seen from this part of the log related to the filtering step, reading the JSON summaries is very fast (notice how the timestamp is the same):
I0629 18:16:41.069377 OrthancPlugins.cpp:1359] Plugin making REST POST call on URI /tools/find (built-in API)
I0629 18:16:41.069377 FilesystemStorage.cpp:154] Reading attachment “2584de72-e00f-4e87-87aa-f497b239a41b” of “JSON summary of DICOM” content type
I0629 18:16:41.069377 FilesystemStorage.cpp:154] Reading attachment “9cede5d9-8d10-42d3-83a6-9ba73e20ce0a” of “JSON summary of DICOM” content type
I0629 18:16:41.069377 FilesystemStorage.cpp:154] Reading attachment “3623ed3d-4188-4373-806e-8b8769ca72f0” of “JSON summary of DICOM” content type
I0629 18:16:41.069377 FilesystemStorage.cpp:154] Reading attachment “999f67e4-249a-41c1-a076-261a4dbc6676” of “JSON summary of DICOM” content type
I0629 18:16:41.069377 FilesystemStorage.cpp:154] Reading attachment “d9d5c99f-36ca-40c3-abe5-9ee813e4b8cd” of “JSON summary of DICOM” content type
I0629 18:16:41.069377 FilesystemStorage.cpp:154] Reading attachment “05f80f11-1f7d-43af-8647-af976d46fac6” of “JSON summary of DICOM” content type
I0629 18:16:41.069377 FilesystemStorage.cpp:154] Reading attachment “d6c12659-d7ae-41ec-bdf6-9d3d54b43672” of “JSON summary of DICOM” content type
I0629 18:16:41.069377 FilesystemStorage.cpp:154] Reading attachment “2c888924-1174-4ef3-9196-bb0c760b9837” of “JSON summary of DICOM” content type
I0629 18:16:41.069377 FilesystemStorage.cpp:154] Reading attachment “ef7704e1-781f-46d7-a222-ca6356a0e4d3” of “JSON summary of DICOM” content type
I0629 18:16:41.069377 FilesystemStorage.cpp:154] Reading attachment “651244bb-25e5-488c-98b3-ba1ab9aa74d9” of “JSON summary of DICOM” content type
So, your problem is not related to a bad performance of your filesystem. However, the calls to “/studies/{…}/instances”, that only make use of your PostgreSQL database, can take long time. For instance, 500ms are needed to access the list of instances of the study “5c0b88d2-c68f035f-b22a0a55-e8ebd753-c2d12245”:
I0629 18:16:46.715725 OrthancPlugins.cpp:1294] Plugin making REST GET call on URI /studies/5c0b88d2-c68f035f-b22a0a55-e8ebd753-c2d12245/instances (built-in API)
I0629 18:16:47.201405 OrthancPlugins.cpp:1294] Plugin making REST GET call on URI /instances/c86455a3-6244f520-1ac5972b-1c72b82f-a8bfb590/tags (built-in API)
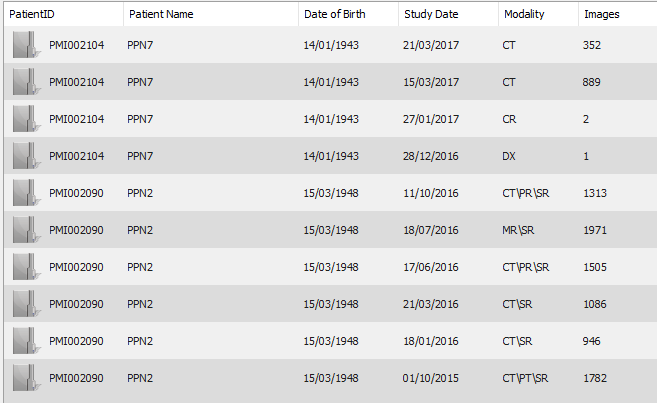
The URI “/studies/{…}/instances” are notably used to populate the “Modality” and “Images” columns of your screenshot. As a consequence, I guess the bad performance you observe is not a bug within the DICOMweb plugin (even if some redundant calls to the REST API of Orthanc could probably be avoided), but instead a direct consequence of the following issue that is related to PostgreSQL:
https://bitbucket.org/sjodogne/orthanc/issues/47/index-improvements-for-pg-plugin
That’s for my analysis. In order to validate it, please could you try to reproduce exactly the same test, with the same database, but without enabling the PostgreSQL plugins? If the slowdown vanishes, your issue is a duplicate of issue #47.
Regards,
Sébastien-