Hello,
I don’t understand why I can’t synchronize two series in the web viewer (see video : https://drive.google.com/file/d/1w3fVLuTYIYlBa8nxtP9zOVIsnp8Ke86u/view?usp=sharing). Is there a bug ? I tried in Firefox and Chrome.
Thanks a lot
David
Hello,
I don’t understand why I can’t synchronize two series in the web viewer (see video : https://drive.google.com/file/d/1w3fVLuTYIYlBa8nxtP9zOVIsnp8Ke86u/view?usp=sharing). Is there a bug ? I tried in Firefox and Chrome.
Thanks a lot
David
You’ve got two different exams - I’m not sure if Osimis support synchronisation of two
Security Clause
Please note that the contents of this message including its attachments is the information protected from disclosure. If this message is not meant for you, please be warned that disclosing, copying, publishing or using it or any attachments is prohibited. If you received this message by mistake, please inform sender immediately, send the message back and delete it from your system including all the attachments.
Right now, only series with exactly the same ImageOrientationPatient can be synchronized. We should maybe have some kind of tolerance there.
Could you check the value of the ImageOrientationPatient in both series and paste them here ?
Hello,
Thanks for your answers, I’m sorry I don’t know how to find the value of the ImageOrientationPatient (I even don’t know what it is).
I understand that synchronization works only for two series in the same exams. But what we’d need is to compare two scans made at different periods to evaluate the evolution of the patient’s lung, so it’s necessarily two different exams (with potentially different number of image, or different orientation). We can do the synchronization with other viewers, that’s why we thought it could be the case with Osimis
I send you the ImageOrientationPatient as soon as I find it
thanks again,
David
Here’s a sample where we compare 2 exams (http://viewer-pro.osimis.io/osimis-viewer/app/index.html?study=b6e8436b-c5835b7b-cecc9576-0483e165-ab5c710b). You may adjust the synchronization by holding the shift-key while using the wheel.
To get the ImageOrientationPatient, open an instance in Orthanc and check for the tags:
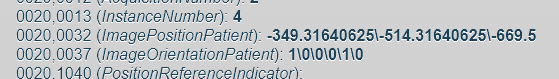
Here are the information :
3 NATIVE THORAX INSPI : 0020,0037 (ImageOrientationPatient): -1\0\0\0\-1\0
Parenchyme THORAX 1.0 Parenchyme : 0020,0037 (ImageOrientationPatient): 1.00000\0.00000\0.00000\0.00000\1.00000\0.00000
The sample you sent is very good, would it be available in the Basic viewer ?
Could you provide me with the 2 anonymized series you wish to synchronize ?
In this case, it seems that the directions are opposite which should be quite easy to handle.
Yes, the synchro feature is working the same way in the basic viewer.
I’d like to send the anonymized series but I don’t know how to anonymize them (I know few things… I’m not a programmer)
You’re right the directions of two scans are opposite (one was done to the front, the other to the back), the user will have to turn the scan
Thanks again
David
Once on a series, just click “anonymize”. It will then open the “Anonymized 1” patient page and click “download zip”
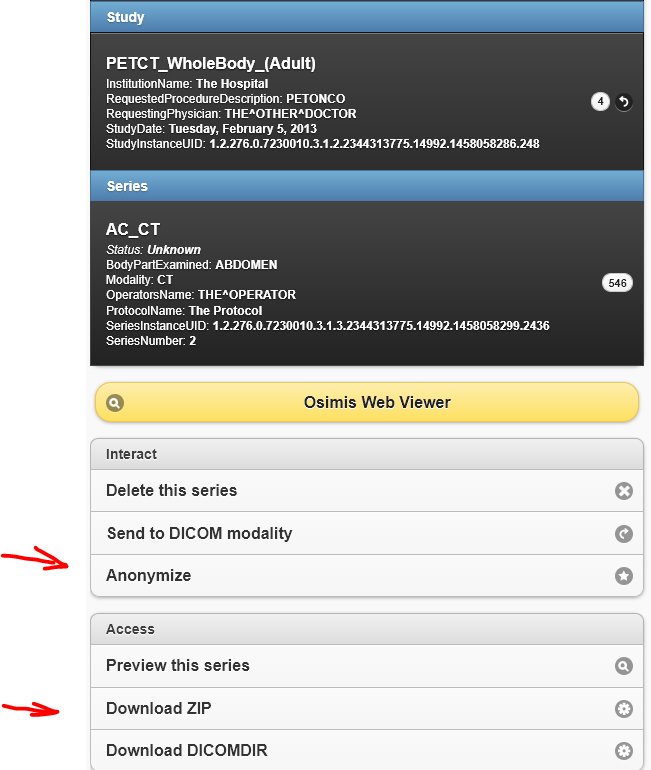
I sent you a link in a personal email to find the two anonymized series
thanks again
David
Hi David,
I’ve fixed the issue in this commit: https://bitbucket.org/osimis/osimis-webviewer-plugin/commits/090cd828a84a754b19b8fec6c8074dece70a8384
Note that, with the sample data you sent me, the Image Position are very distant between the two series so you’ll have to first disable the synchro (by pressing “shift”), select a slice in each series then enable synchro again (by releasing “shift”).
A windows version is available here: orthanc.osimis.io/win64/viewer/090cd82/OsimisWebViewer.dll
If you’re using Docker, that will be part of a future release of osimis/orthanc images.
Best regards,
Alain.
Hi, it’s ok for us to change the synchro by pressing “shift”. I can’t test it because I don’t use Windows, do you know when the next release is ?
thanks a lot
best regards
David
I’ve just triggered a build of osimis/orthanc:18.9.2.
You’ll need to define WVB_ALPHA=true in order to activate the alpha version. (https://osimis.atlassian.net/wiki/spaces/OKB/pages/26738689/How+to+use+osimis+orthanc+Docker+images#Howtouseosimis/orthancDockerimages?-OsimisWebViewer)
is this a new feature in Osimis viewer ?
It’s there since the 1.1.0 release: https://bitbucket.org/osimis/osimis-webviewer-plugin/src/master/RELEASE_NOTES.txt
Hello Alain, hello all,
The synchronization works for two different exams, but I detected a strange thing you can see on this video : https://drive.google.com/file/d/1ZmR4wOj1giRErDlNoRld2J-Vj7zjbCH4/view?usp=sharing
When I add a new serie, the synchronization doesn’t works well (I can’t see the two series in parallel, and when I click “play” it always go back to the beginning of the serie). But when I de-synchronize, click play, and then synchronize again, it works well. Can you see it ?
best regards
David
Hi David,
What you observe is exactly what I described in my previous email:
Note that, with the sample data you sent me, the Image Position are very distant between the two series so you’ll have to first disable the synchro (by pressing “shift”), select a slice in each series then enable synchro again (by releasing “shift”).
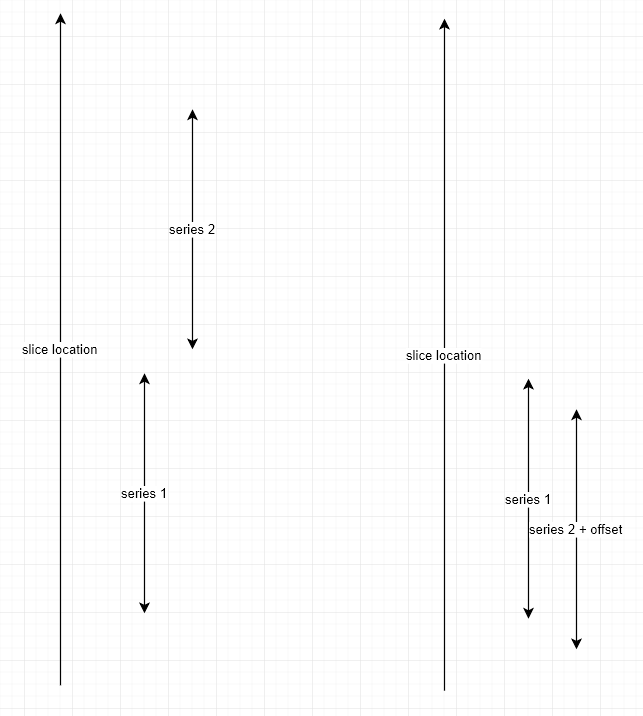
The locations of your slices (according to the Dicom tags) are explained by the left diagram below.
If you click on a slice in series 1, there’s no corresponding slice in series 2 so we display a black image. Same happens when you click on series 2.
Once you press shift, you release the synchro so you can adjust the displayed slices in each series. On you release the shift key, the sync tool will record the offset between the slices and apply it later on such that series 1 and series 2 seems to be at the same locations (as shown in the right part of the diagram).
Ok, thank you for your answer, I must admit that I don’t understand everything, but in other viewers we use (ex: Horos), there is no black image when we scroll (the 2 series begin to be scrolled even if they are not sychronized). The fonctioning of Osimis will certainly seem strange for our users.
best regards
David