Dear John,
Sorry for the delay.
That setup has worked for several months until recently when I had to switch from calling “/studies/{ID}/anonymize” to calling on an instance level, “instances/{ID}/anonymize”.
Yes, the anonymization and modification primitives work differently at the instance level, than at the patient/study/series levels. As described in the Orthanc Book, anonymizing/modifying one single instance will return the anonymized/modifed instance in the body of the REST answer (whereas when applied at the patient/study/series levels, the result is stored back in Orthanc):
http://book.orthanc-server.com/users/anonymization.html#modification-of-a-single-instance
My question is, where is the AnonymizedFrom meta stored? Or set for that matter?
It is set by the Orthanc core, using the metadata primitive. Metadata consists in an associative key-value array (mapping a integer key in the range [0,65535] to a string value) that is associated with each DICOM resource stored inside Orthanc (may it be a patient, a study, a series or an instance). Metadata records information that is not readily available in the DICOM tags. “AnonymizedFrom” is one such metadata (whose integer key is 6).
Metadata associated associated with one instance “id” can be accessed through the REST API:
When you modify/anonymize a single instance, the result is not stored inside the Orthanc database, so “AnonymizedFrom” is unavailable.
Should a call to anonymize at the instance level be setting the AnonymizedFrom field or do I need to do that myself? And if I need to set this meta data, how do I do it?
As “AnonymizedFrom” and “ModifiedFrom” are handled privately by Orthanc, they are read-only metadata. You cannot set these fields by yourself.
However, you can mimic their behavior by using user-defined metadata. Such metadata is associated with an integer key greater or equal to 1024. You can associate a symbolic name to user-defined metadata using the “UserMetadata” configuration option of Orthanc. For instance, here is how you would set/get metadata 1024:
hello
I should add that because I am running anonymize at the instance level, I am creating new Study, Series and Instance UID manually before calling anonymize and then feeding them to anonymize via the “replace” option. I do not let anonymize generate UID itself. Maybe anonymize ONLY sets AnonymizedFrom when it handles the UID generation? And I’ve skipped that step by generating UID myself?
You are free of generating UIDs by yourself. The metadata AnonymizedFrom is unrelated to this.
I found the routine AnonymizeOrModifyResource in OrthancRestAnonymizeModify and see the code where the metadata is set up to point from anonymized/modified files to original files.
Checking the metadata table in the PostGres database I can see that my newly anonymized/modified Dicom are missing these metadata connections.
Metadata are stored in the table called “metadata”. Here is a screenshot of pgadmin3 showing how the AnonymizedFrom metadata (type 6) is associated to the original identifier:
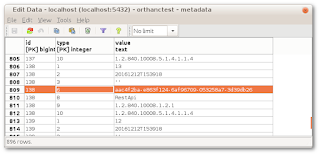
Perhaps that data is never updated when anonymizing at the instance level? After all, unlike anonymizing at the Patient/Study/Series levels, which generate new Patient/Study/Series within Orthanc itself and return metadata, a call to /instances/{ID}/anonymize will RETURN the anonymized DICOM itself. This must then be uploaded back into Orthanc, if you want it there.
Yes, that’s exactly the point: Metadata are only available for resources stored inside Orthanc. Once one instance leaves the Orthanc ecosystem (e.g. through anonymization), metadata is lost.
Is there a way to call anonymize at the instance level and have Orthanc keep a record in the metadata table of the links to the original DICOM?
The “AnonymizedFrom” is read-only, so you cannot set it by yourself. However, you could set an user-defined metadata.
Similarly, is there a way to have it simply retain the new DICOM, rather than sending it in response.
No, this is not possible in the REST API.
That being explained, you have at least 2 solutions to your issue:
- As Orthanc is lightweight, you can use 2 instances of Orthanc. The first one is responsible for receiving files, anonymizing individual instances, then sending the anonymized instance to another Orthanc server (possibly located on the same host). You then know that each instance stored by the second Orthanc server only contains anonymized instances, which allows to avoid infinite loops in your scripts.
- The “anonymize” URI of Orthanc automatically sets the (0012,0063) “DeidentificationMethod” DICOM tag. On receiving a new instance, your Lua script could check whether this tag is set to know whether the instance results from an anonymization or not.
The “best” solution clearly depends upon your application.
HTH,
Sébastien-
